Our DEL-AI discovery engine leverages the combined power of our data-rich DEL capabilities, automated chemistry, and machine learning to accelerate drug discovery and unlock a whole new universe of opportunity for patient treatment.
Data-enriched drug development
Targeted protein degradation is an unprecedented therapeutic approach that harnesses the cell’s natural protein degradation machinery to eliminate disease-causing proteins. This field merges new biology and chemistry, demanding extensive research and new levels of analysis of big data. To navigate this new paradigm in drug discovery, Nurix has adopted an empirical approach implemented on a truly grand scale. Our high-throughput data generation workflows form the backbone of our research platform. Through our investment in technologies like DNA-encoded libraries (DEL), automated chemistry, and direct-to-biology screening, Nurix emphasizes a data-driven approach across our research pipeline. Consequently, we have amassed one of the largest datasets specifically generated from targeted protein degradation research.
DEL-AI discovery engine
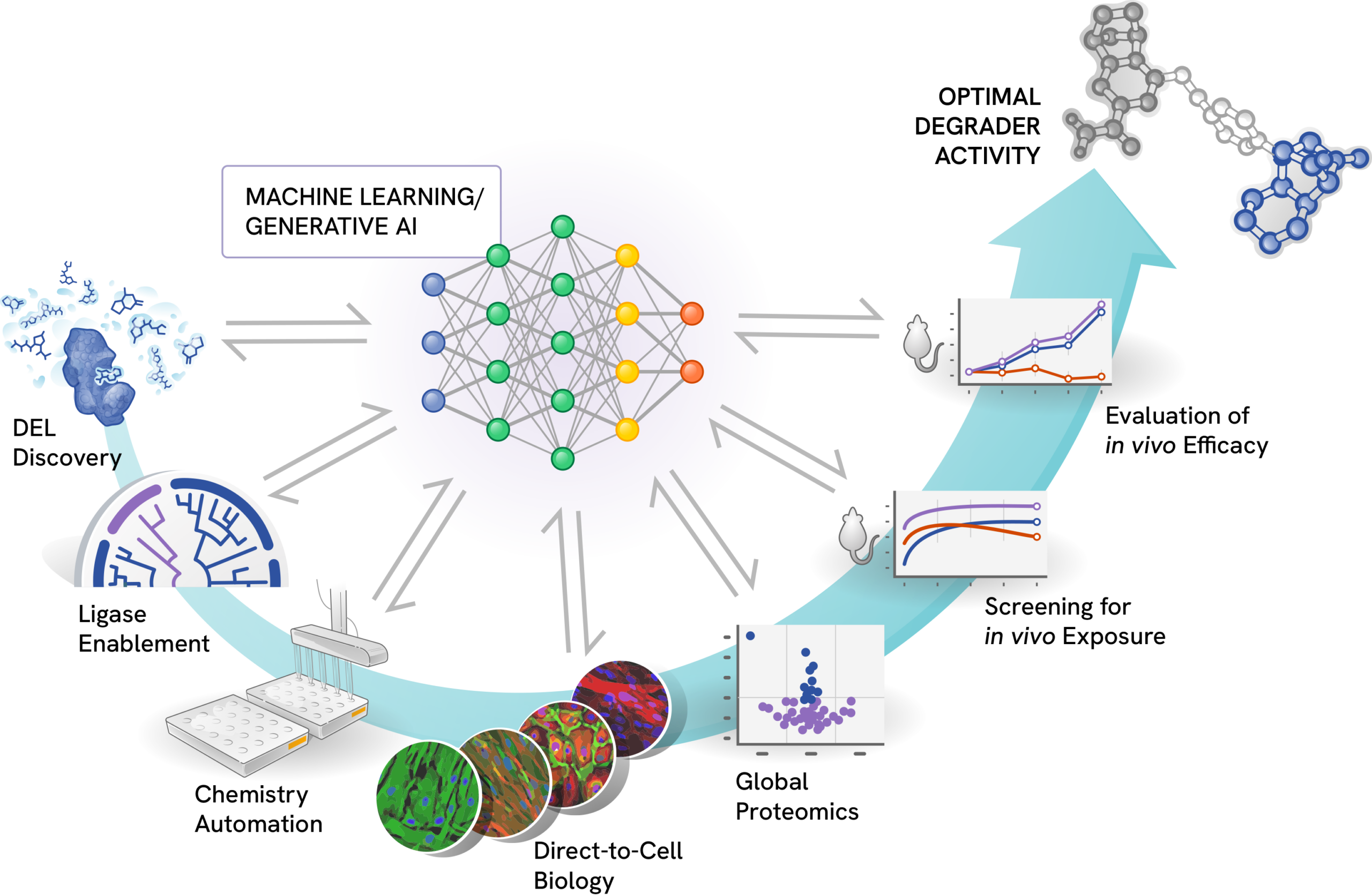
DEL-AI harnesses Nurix’s varied high-throughput data streams to learn the rulebook of targeted protein degradation, delivering a powerful research engine that drives our expanding portfolio of drug candidates. By integrating data across our degrader development activities with deep learning and generative design, Nurix researchers leverage DEL-AI to identify and deliver novel, optimized compounds at any stage along the research pipeline.
DEL Discovery
At the core of our drug discovery process, our DEL technology is designed to scale seamlessly with the demands of machine learning:
- Vast chemical space: Over 5 billion unique chemical structures enable exploration of an unprecedented range of binders for target proteins and E3 ligases.
- Customized scaffolds: Proprietary scaffold-based DELs provide unique geometries and high sp3 character, identifying binders for challenging protein surfaces.
- Rapid hit follow-up: The combinatorial nature of our DELs facilitates quick optimization of promising hits.

Ligase Enablement
An E3 ligase is an enzyme that plays a crucial role in the ubiquitin-proteasome system, a cellular mechanism that ensures that damaged, misfolded, or unneeded proteins are broken down and recycled. The human genome encodes over 600 E3 ligases, each with a specific function. Historically, E3 ligases were considered undruggable, and until recently, medical research focused primarily on two, well-understood E3 ligases: cereblon and VHL. We have enabled over 90 E3 ligases in our drug discovery process. Our expertise in the structure and function of E3 ligases allowed us to create DNA-encoded libraries specifically designed to identify drugs that harness or inhibit E3 ligases.
Chemistry Automation
Our chemistry platform can rapidly synthesize and purify hundreds of compounds, enabling quick iteration and optimization of lead compounds. By creating more molecules and empirically testing their properties, Nurix is rapidly learning the rulebook of degradation to enable smarter drug discovery and development.
High-throughput Biology
We employ a direct-to-biology approach with automated assays to evaluate compounds for target engagement, cellular activity, and drug-like properties, allowing us to test numerous compounds from our automated chemistry in biologic assays of protein degradation and in vivo pharmacology.